Applications and Collaborations
Ongoing applications of Force Field X include the following collaborations:
- Understanding missense variants in the context of inherited deafness.
- Creation of an alchemical pipeline to predict the crystallization of pharmaceuticals.
- Predicting the binding affinity between cyclodextrin hosts and guest ligands.
Understanding Missense Variants in the Context of Inherited Deafness
| The Deafness Variation Database (DVD) provides a comprehensive guide to genetic variation in genes known to be associated with deafness. The DVD v9 includes all known genetic variants present in 223 deafness-associated genes that are included on OtoSCOPE®, a comprehensive genetic deafness screening platform. For each of these genes that codes for a protein, Force Field X was used to refine an AlphaFold2 structures using the AMOEBA force field and our novel global optimization algorithms. These structures are more consistent with the chemical features of protein structures from X-ray crystallography as assessed by the tool MolProbity. |
|
The protein models are available in the DVD -- please check them out here! |
|
For more information, please see the following publication from our collaboration with labs of Richard J. H. Smith, Thomas Casavant and Terry Braun: Tollefson, M. R., Gogal, R. A., Weaver, A. M., Schaefer, A. M., Marini, R. J., Azaiez, H., Kolbe, D. L., Wang, D., Weaver, A. E., Casavant, T. L., Braun, T. A., Smith, R. J. H., and Schnieders, M. J., Assessing Variants of Uncertain Significance Implicated in Hearing Loss Using a Comprehensive Deafness Proteome. Human Genetics 2023, 142, 819–834. |
Alchemical Polymorph Prediction Pipeline
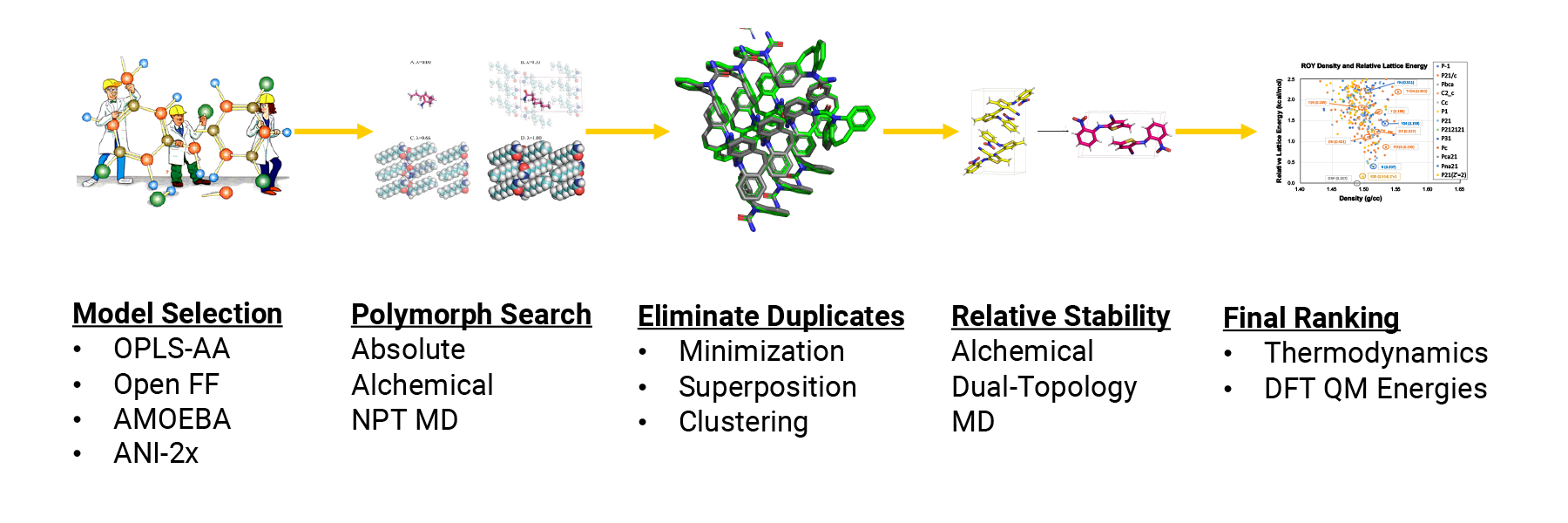
| We are collaborating with Mitsubishi Tanabe Pharma and Takeda to create a complete crystal structure prediction pipeline using Force Field X. |
|
|
Please see our recent joint publication describing the "Progressive Alignment of Crystals" (PAC) algorithm: Nessler, A. J., Okada, O., Hermon, M. J., Nagata, H., and Schnieders, M. J., Progressive Alignment of Crystals: Reproducible and Efficient Assessment of Crystal Structure Similarity. Journal of Applied Crystallography 2022, 55, 1528-1537. |
Cyclodextrin Host-Guest Binding Affinity
| This is a new collaboration with Pfizer -- more soon on our progress. |
|