Welcome to Force Field X
Introduction
|
Force Field X is an open source polyglot software for molecular biophysics that targets open research questions in the areas of:
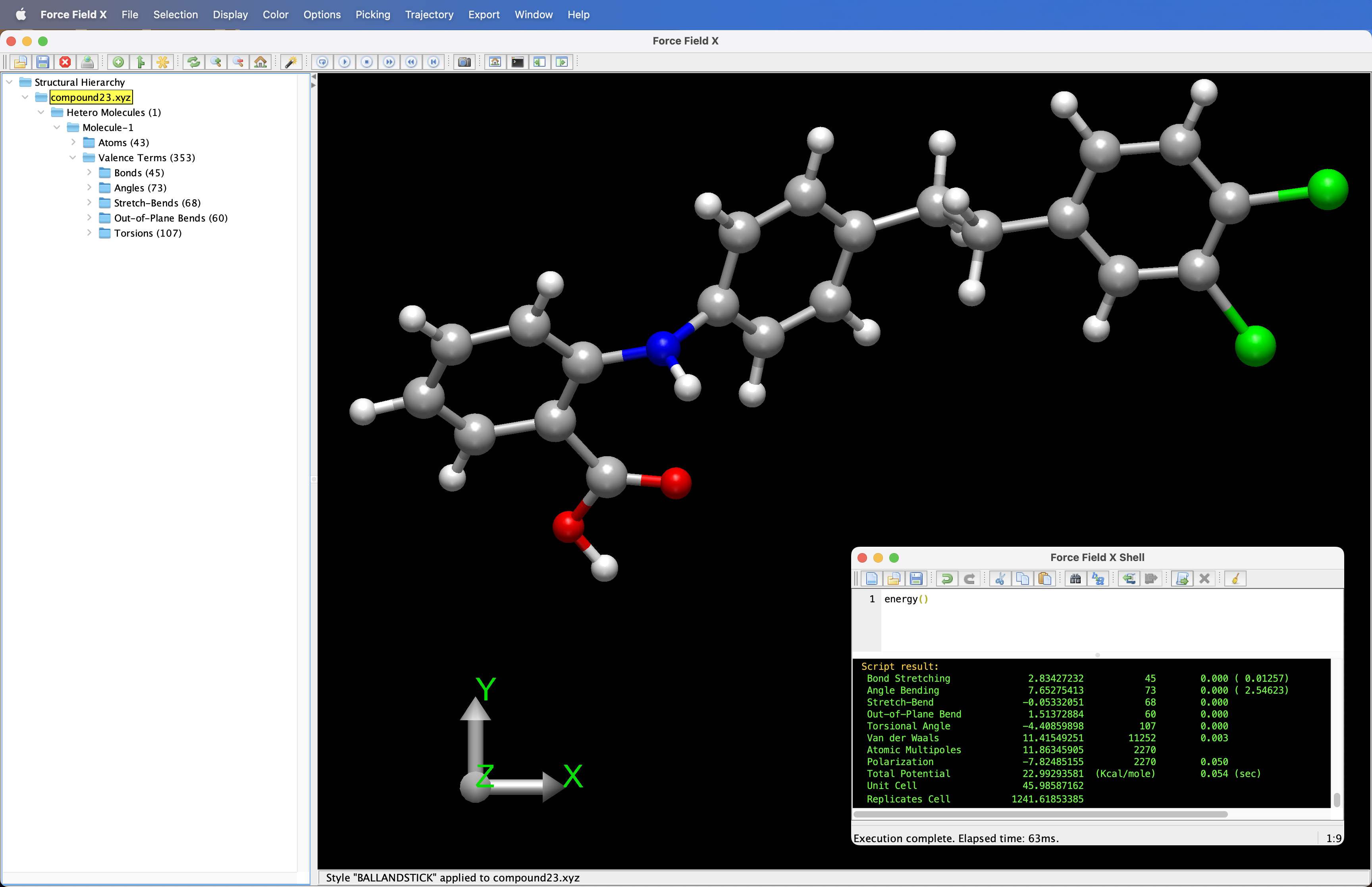
The graphical user interface (GUI) screenshot to the right shows "Compound 23", which was the 23rd molecule predicted as part of the CCDC crystal structure prediction series. Performance
|
|
Getting Started
Instructions on how to download, install and use Force Field X are available in the following sections.
|
Download |
Install |
User Manual |
Applications |
|
Method Development |
Build from Source |
Programmer Guide |
Publications |
Funding Support
Development of Force Field X has been supported by the following organizations.
|
|
|
|
|
| NSF CHE-1751688 | NIH R01 DC012049 | SFARI Award 1019623 | Mid-Career Scholar Award |